Quantitative PCR
Contributors
Students
- Jaroslaw Ciba, Department of Physics, creates visual designs
- Thomas Travis, Department of Physics, oversees interactive aspects of design
- Marcus Essam, Department of Medicine, in charge of overall design
- Jacky Zhu, Department of Medicine, edits and updates wiki page
Staff Partners
- Anabel Varela Carver, Department of Surgery & Cancer. Staff partner from October 2021.
- Joana Dos Santos, Department of Surgery & Cancer. Staff partner from October 2021.
- Jon Krell, Department of Surgery & Cancer. Staff partner from October 2021.
- Robert Kypta, Department of Surgery & Cancer. Staff partner from October 2021.
Aims & Learning Outcomes
Intended Use and Audience
This visualization is intended for self-study to support iBSc Cancer Frontiers Module 1 teaching (q-PCR principles, data analysis and application to an experiment involving cancer cells)
Learners are expected to have an basic understanding of how PCR functions as well as the purposes of using this technology, which are part of the standard A-level Biology curriculum.
Topic Background
qPCR is a laboratory technique widely employed in life science related subjects. It is build upon the foundation of PCR, a technological innovation allowing exponential amplification of genetic material. qPCR (Quantifying PCR) builds directly upon this concept, while enabling researchers to monitor the quantity of genetic material in real time as they amplify, hence the name qPCR or real-time qPCR. Through the analysis of data generated by qPCR, researchers will be able to conclude the presence or absence of a gene inside samples, as well as comparing their amounts in relative terms. Its most recent usage that gained global attention was its contributions to combating COVID-19 through viral DNA testing, which is considered the most accurate test to discover a current infection even in asymptomatic individuals.
Intended Learning Outcome
After using this visualization, students should be able to explain how changes in Ct values from a q-PCR experiment relate to changes in the numbers of copies of a gene that a cell contains (for example, a cancer cell upon treatment with a drug).
- Understand how PCR is applied to various biological research
- Describe the workflow of PCR centered protocols
- Explain the roles of different reagents used in PCR
- Provide basic analysis based on qPCR data, such as CT values
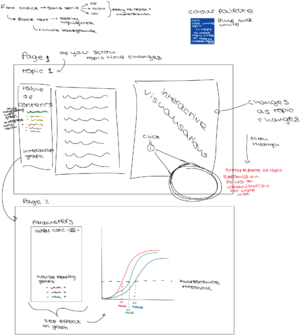
Design Overview
The overall flow of the visualization are divided into two parts:
- Explanation of the basic principles of PCR, including necessary reagents and what happens within one thermal cycle.
- Explanation of how to interpret PCR results (CT value in particular), as well as what factors may influence the result.
Each of these parts will be explored with multiple slides aiming to introduce material to students in manageable bits. For instance, PCR principles will be dissected down into lab flow, protocols, reagents, etc., whereas PCR results will be explored by taking apart separate data for examination first before bringing them together to form the bigger picture.
For a view of the entire project so far: https://imperiallondon-my.sharepoint.com/:p:/g/personal/jwc20_ic_ac_uk/EfWqmugEj01NlH_zrMdI9TgB94U2218wx4Q-P2nf1poV_Q?e=i9euEO&isSPOFile=1
The Thermal Cycle Part has this animation in mind: https://imperiallondon-my.sharepoint.com/:p:/g/personal/jwc20_ic_ac_uk/EZAbz8uaTLxLtdJ3uAYK704BnMIbwp2jZ6ynFvNlL0tkQw?e=0v3Zgi
Design Justification
Assessment Criteria
Education Design
- Target audience and their prior knowledge is clearly identified. This information is applied to decide what the learning outcomes should be.
- Key concepts identified and broken down into several discrete and easily achievable points.
Graphical Design
- The design is such that the visualization is clear and easily understood; the layout isn't cluttered.
- The choice of font, as well as size and placement of text, makes the information easy to comprehend.
- The color palette is appropriate and well justified, making it easy to see all text/design elements
Interaction Design
- The learner should know intuitively what the objectives are of the visualization.
- The user should be able to intuitively understand the function of each interactive element in the visualization.
- There should be immediate visual feedback when a learner interacts with the visualization.
Education Design
- Diagrams - easy to read - convey a lot of information in understandable sections - can use lots of colour and make certain areas stand out
- Decided to remove the text box and go with diagrams with small bitesize amounts of information
- Use of imperial colour scheme (dark blue yellows, whites) high contrast colours so easy to see and clear and suggested by peers during initial peer reviews of initial draft design
Graphical Design
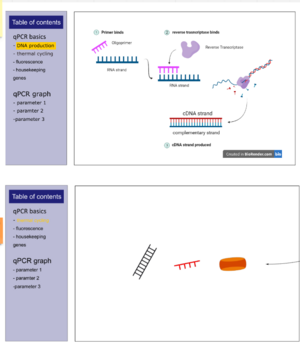
- Most spaces are filled with custom made diagrams with captions to explain the different elements involved.
- Diagrams are designed or borrowed from biorender.com
- Pages have a side section explaining concepts in text and a graphical panel on the right
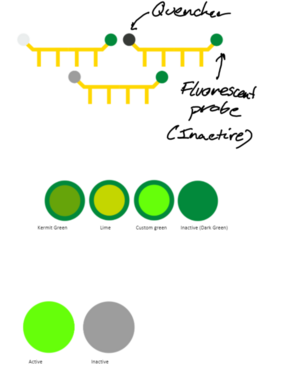
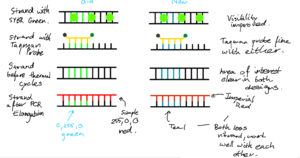
- Taqman Probe Design - The body of the probe was made in a style consistent with previous visuals - sharp angles, no rounding, mostly straight lines. - Lemon yellow (#FFDD00) used for the body of the probe, from the Imperial palette. Differentiates the probe from other elements;different colour from thermal cycles elements; - Quencher. Completely passive element, so grey used to not stand out. Different greys from the Imperial palette looked at, and finally settled on dark grey (#373A36). Light grey a bit difficult to see against the background, whilst dark grey simply looks better than cool grey. - The fluorescent probe emits green light; Imperials dark green used to show that the probe is inactive. When probe is detached off taqman, fluorescene starts. The greens featured on the Imperial colour palette not deemed suitable. A more vibrant light green used (#66ff0a) after playing around with the colours.
- SYBR Green Design - An inactive and active version exist; when attached to dsDNA, the dye is active. Inactive otherwise. This is simply shown via colour change. - Inactive version uses cool grey. Dark grey would have been used, however as this was already used for the quencher, we do not want to confuse the learner, so different colour chosen. - Active version features the same lime green that Taqman used, to indicate fluorescence consistently between the two. Note: Perhaps will switch cool grey and dark grey, as cool grey actually looks better when taqman probe is attached to strands.
- DNA Strands - Chose for a simple design made of straight lines. A lot of abstraction is allowed, as learners focus is on understanding PCR/qPCR - they already have brilliant grasps on the structure of DNA and other reagants, so do not need the detail. - For example, the double helix of structure of DNA is simply reduced to straight lines with bases connecting them. Bases in uniform colour too - we do not need to match complementary bases, learners are more than aware of it! We instead use different colours to signfify areas of interest on the strands. Specifically, a simple green (0,255,0) was used to indicate area where the primer attaches, making it stand out. Later, this was changed to Imperials Teal for consistency with the Imperial palette and for contrast with SYBR green, reserving green for fluorescence. - Different colours for DNA were trialled, with black eventually chosen for simplicity. - The thickness of strands was also played around with. Very thin and very thick designs both look awful, and a midrange thickness was chosen. - Rounded caps are used at the ends of the strands as they do look better.
- Polymerase - Polymerase simplified to a rounded rectangle, with two ellipses, similar to some designs in research. - The rectangle is orange from the imperial palette (#D24000), whilst the ellipses are tangerine (#EC7300). The darker colour is used to indicate depth, as the elliptical components of the polymerase "wrap" around a strand.
- Primer - Settled on red for contrasting against the green of area of interest on DNA strands. - Imperials Red used (#DD2501) in final design, for consistency with the Imperial palette.
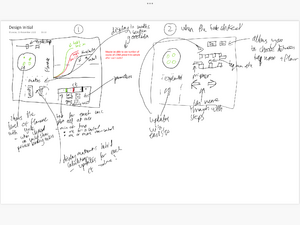
Interaction Design
- Our first design included a large text box section however with feedback and self reflection we realised this wouldn't be engaging for the user even though it would be an efficient way to display information in a small space
- One page will allow users to tweak with PCR parameters to observe how it can affect experimental outcomes
- In most part of the visualization at this stage though, the interaction is limited to PPT style animation presentation
Progress and Future Work
- The design is closed to being finalized, with certain tweaks still on the way
- Updated project designs will be continuously uploaded to this wiki page
- Programming still need work in order for certain features of our design to be feasible
IMPORTANT: This wiki page is still a work in progress. While the overall structure is unlikely to change, certain elements have not been completely filled in yet, including the different aspects of design justification (above section).
Links
- Link to GitHub repository for code in development:
- Link to visualisation on ImpVis website (when uploaded):
- Link to Collection on ImpVis website (when created):
- https://miro.com/app/board/o9J_lpvA2AE=/?invite_link_id=762082753954